Boltz-2 Protein Structure Prediction
Predicts 3D protein structures from MSA (A3M) and FASTA using the Boltz-2 model. Supports batch prediction, confidence scoring, and reproducible sampling.
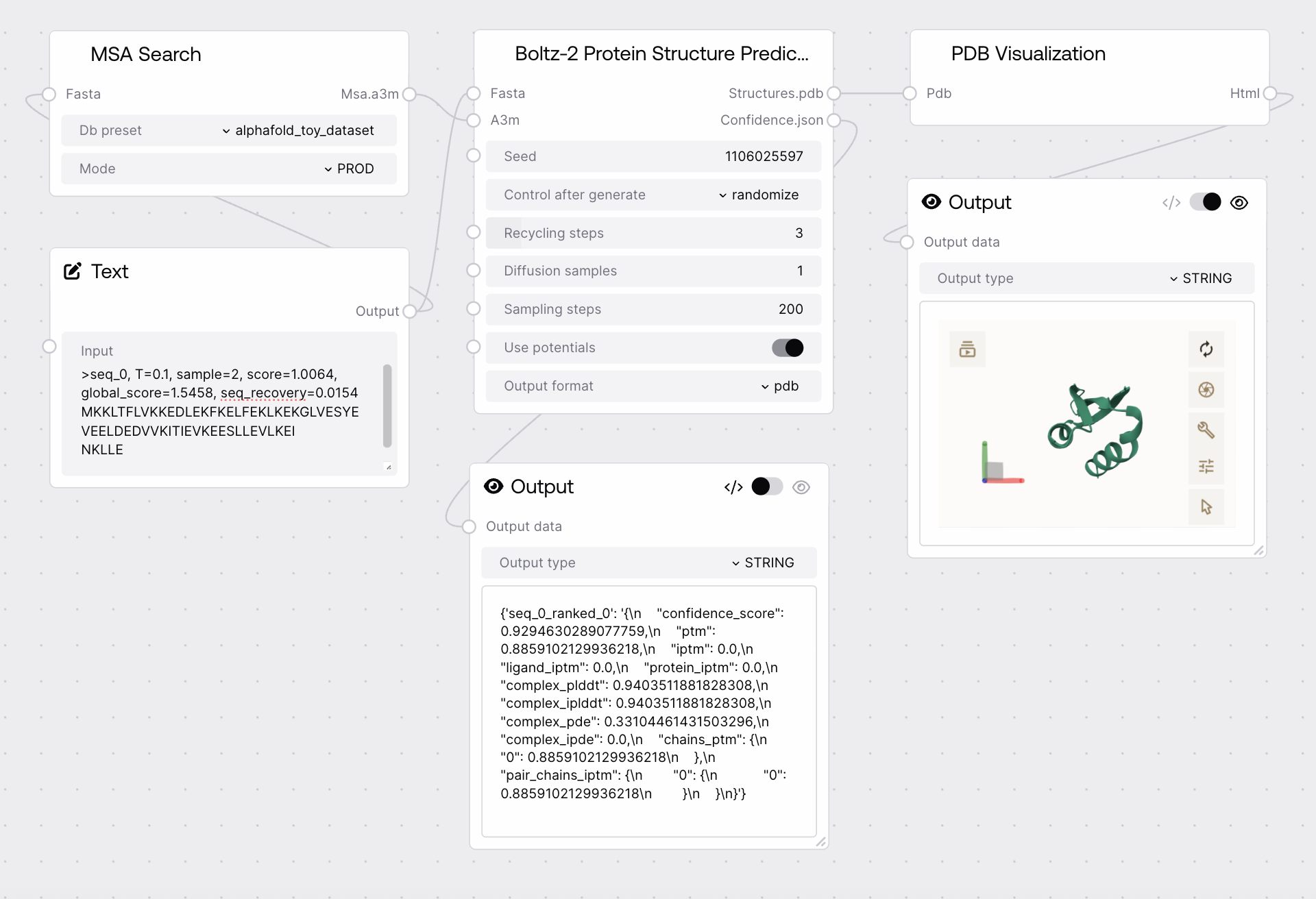
Quick Start
- Prepare your protein FASTA sequence and MSA (A3M format).
- Set the desired number of structure samples and recycling steps.
- Run the node to generate ranked structure predictions and confidence scores.
Setup Guide
- Obtain the target protein's FASTA sequence.
- Run MSA search to generate A3M input (e.g., with MSA Search node).
- Set
recycling_steps for quality vs. speed.
- Choose
diffusion_samples for diversity.
- Optionally adjust
sampling_steps, use_potentials, and output_format.
3. Run and Retrieve Results
- Execute the node.
- Download predicted structures and confidence scores from outputs.
Basic Usage
Single Protein Structure Prediction
- Input a single FASTA and A3M to predict structure.
- Adjust
recycling_steps for accuracy.
- Use
diffusion_samples >1 for diverse predictions.
Batch Prediction
- Provide A3M with multiple entries to predict structures for each sequence.
- Each output is indexed by input sequence ID.
Configuration
| Field |
Description |
Type |
Example |
| fasta |
Original protein FASTA sequence |
FASTA |
">A\nMKT..." |
| a3m |
MSA search results (A3M format) |
A3M |
{"A": "..."} |
| seed |
Random seed for reproducibility |
INT |
42 |
| Field |
Description |
Type |
Example |
| recycling_steps |
Number of recycling iterations |
INT |
3 |
| diffusion_samples |
Number of structure samples to generate |
INT |
5 |
| sampling_steps |
Steps for diffusion process |
INT |
200 |
| use_potentials |
Use potentials for higher quality |
BOOLEAN |
True |
| output_format |
Output file format ("pdb" or "mmcif") |
STRING |
"pdb" |
Outputs
| Field |
Description |
Example |
| structures.pdb |
Ranked predicted structures (dict) |
{"A_rank_0": "..."} |
| confidence.json |
Confidence scores for each prediction |
{"A_rank_0": 0.92} |
Best Practices
- Use high-quality, full-length FASTA sequences.
- Generate A3M using comprehensive MSA search for best results.
Parameter Tuning
- Increase
recycling_steps for higher accuracy (at cost of speed).
- Use
diffusion_samples >1 to explore structural diversity.
Troubleshooting
Common Issues
- Missing or empty FASTA/A3M: Ensure both inputs are provided and correctly formatted.
- Low confidence scores: Try increasing
recycling_steps or improving MSA quality.
- Output is empty: Check input sequence IDs and formats.
Need Help?
- Contact support for further assistance.
