Alphafold¶
Runs Alphafold model inference on MSA search results and generates 5 pdb foldings per input sample ranked by best score.
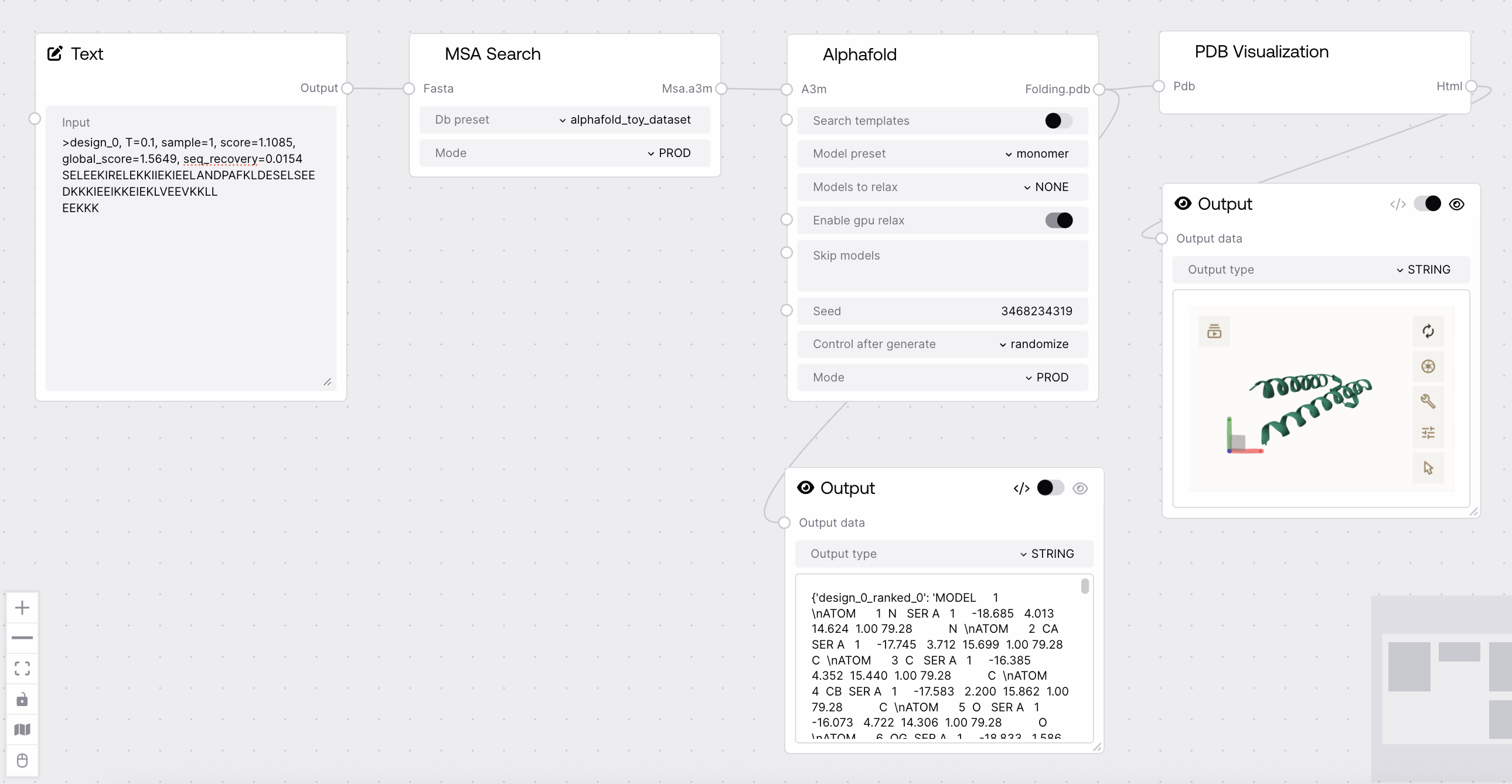
Quick Start¶
- Prepare MSA search results in A3M format.
- Select model and configuration options.
- Run the node to generate ranked protein structure predictions.
Setup Guide¶
1. Prepare Input Data¶
- Obtain MSA search results in A3M format (e.g., using the MSA Search node).
- Ensure input IDs are unique and correspond to your samples.
2. Configure Alphafold Node¶
- Choose model preset and relaxation options as needed.
- Set additional parameters (e.g., skip models, seed).
3. Run Prediction¶
- Execute the node to obtain ranked PDB foldings for each input.
Basic Usage¶
Single Sequence Prediction¶
- Input a single A3M record to predict its structure.
- Use default model and settings for standard monomer prediction.
Batch Prediction¶
- Input multiple A3M records for batch structure prediction.
- Each input will yield 5 ranked PDB outputs.
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| a3m | MSA search results in a3m format. | A3M | {"sample1": "..."} |
| search_templates | Whether to search for available templates before prediction. | BOOLEAN | False |
| model_preset | Which Alphafold model to run. | COMBO | monomer |
| models_to_relax | Whether to run relaxation step (not supported yet). | COMBO | NONE |
| enable_gpu_relax | Whether to run relaxation step on GPU or CPU. | BOOLEAN | True |
| skip_models | Which models to skip (comma-separated indices 1-5). | STRING | 3,5 |
| seed | Base seed for randomness. | INT | 42 |
| mode | Mode to run the node in (MOCK, PROD, TEST). | COMBO | PROD |
Optional Inputs¶
None
Outputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| folding.pdb | Ranked foldings as a dict {pdb_id_rank_id: pdb_content}. | PDB | {"sample1_ranked_0": "..."} |
Best Practices¶
Input Preparation¶
- Ensure A3M input records are correctly formatted and IDs are unique.
- Use the MSA Search node for compatible input generation.
Model Selection¶
- Use the default monomer model for standard predictions.
- Only skip models if you need to speed up inference and understand the implications.
Troubleshooting¶
Common Issues¶
- All models skipped: Cannot skip all 5 models; remove at least one from
skip_models. - Relaxation not supported: Relaxation step is not implemented; set
models_to_relaxto NONE. - Multimer not supported: Multimer model is not available; use monomer presets.
- Input ID mismatch: Ensure input IDs in A3M and FASTA match exactly.
Need Help?¶
- Alphafold documentation
- Contact support for further assistance.