RFDiffusionContigmapConfig¶
Provides interface for passing contigmap configuration to RF Diffusion node.
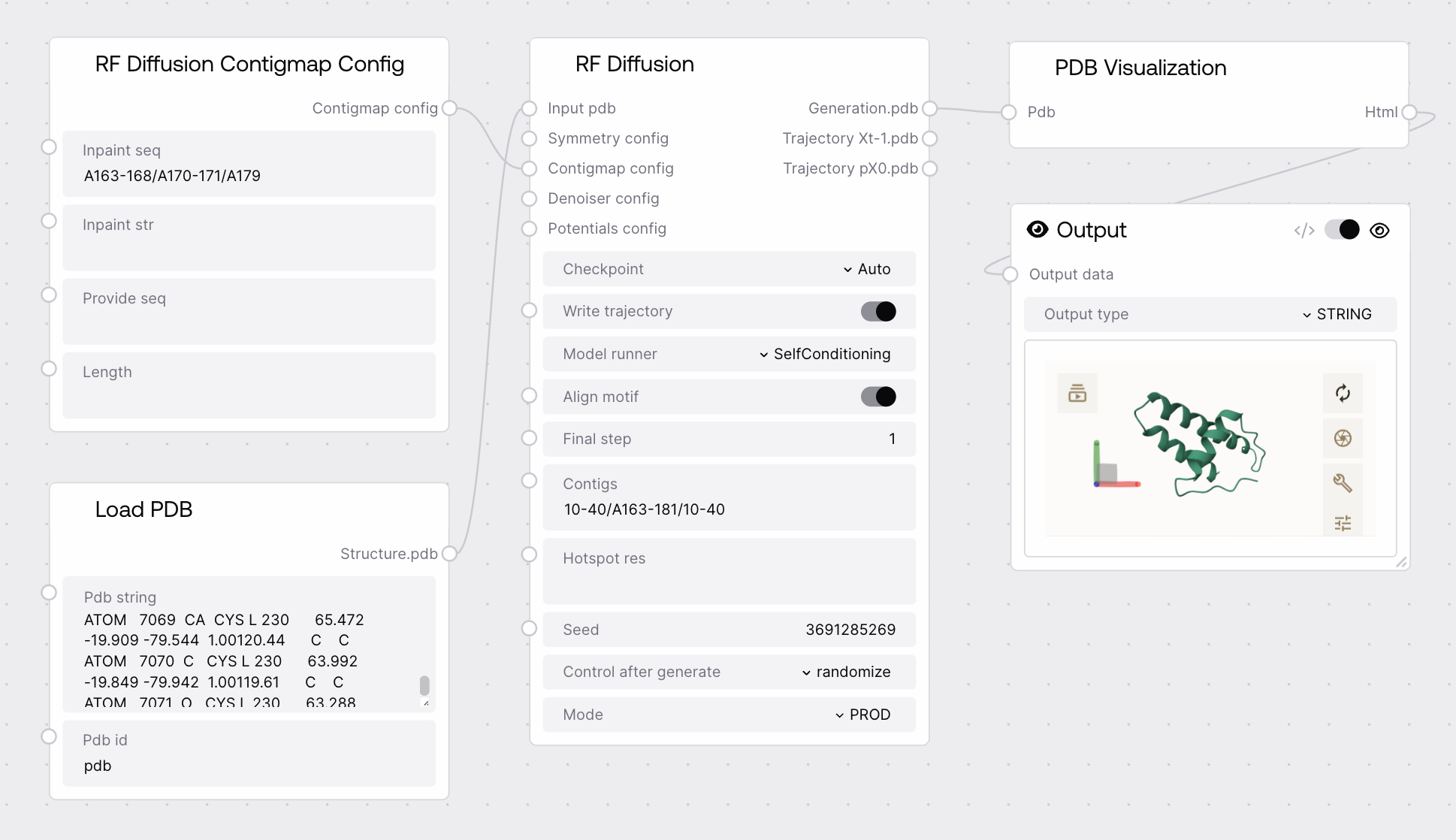
Quick Start¶
- Add the RFDiffusionContigmapConfig node to your workflow.
- Specify masking and unmasking regions as needed for your design.
- Connect the output to the RF Diffusion node's
contigmap_configinput.
Setup Guide¶
1. Add Node to Workflow¶
- Insert the RFDiffusionContigmapConfig node from the node palette.
- Connect it to the
contigmap_configinput of the RF Diffusion node.
2. Configure Parameters¶
- Set masking and unmasking regions using the input fields:
- Use the same format as for contigs (e.g.,
A10-25,5-15). - Leave fields empty if not required.
- Use the same format as for contigs (e.g.,
Basic Usage¶
Masking Sequence Regions¶
- Use
inpaint_seqto mask specific amino acid sequence regions. - Use
inpaint_strto mask 3D structure indices. - Use
provide_seqto unmask specific sequence regions. - Use
lengthto define the length or range for the region.
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| inpaint_seq | Amino acids whose sequence should be masked (contigs format). | STRING | A10-25 |
| inpaint_str | Indices for masking the 3D structure. | STRING | B165-178 |
| provide_seq | Inclusive ranges of sequence regions to unmask. | STRING | 172-177,200-205 |
| length | Length or range of sequence length. | STRING | 100 or 100-150 |
Optional Inputs¶
None
Outputs¶
| Field | Description | Example |
|---|---|---|
| contigmap_config | Configuration dict for RF Diffusion node. | {"inpaint_seq": ["A10-25"], ...} |
Best Practices¶
Parameter Formatting¶
- Use the same format as for RF Diffusion contigs (e.g.,
A10-25,5-15). - Leave fields empty if not needed for your use case.
Workflow Integration¶
- Always connect this node to the
contigmap_configinput of the RF Diffusion node for advanced masking/unmasking.
Troubleshooting¶
Common Issues¶
- Incorrect format in input fields: Ensure you use the correct contigs format (e.g.,
A10-25). - Unexpected results in RF Diffusion: Double-check that masking/unmasking regions are specified as intended.
Need Help?¶
- See the RF Diffusion documentation for more details on contigmap configuration.
- Contact support for further assistance.