RF Diffusion
Runs inference of a RF Diffusion model (conditional or unconditional) for protein structure generation.
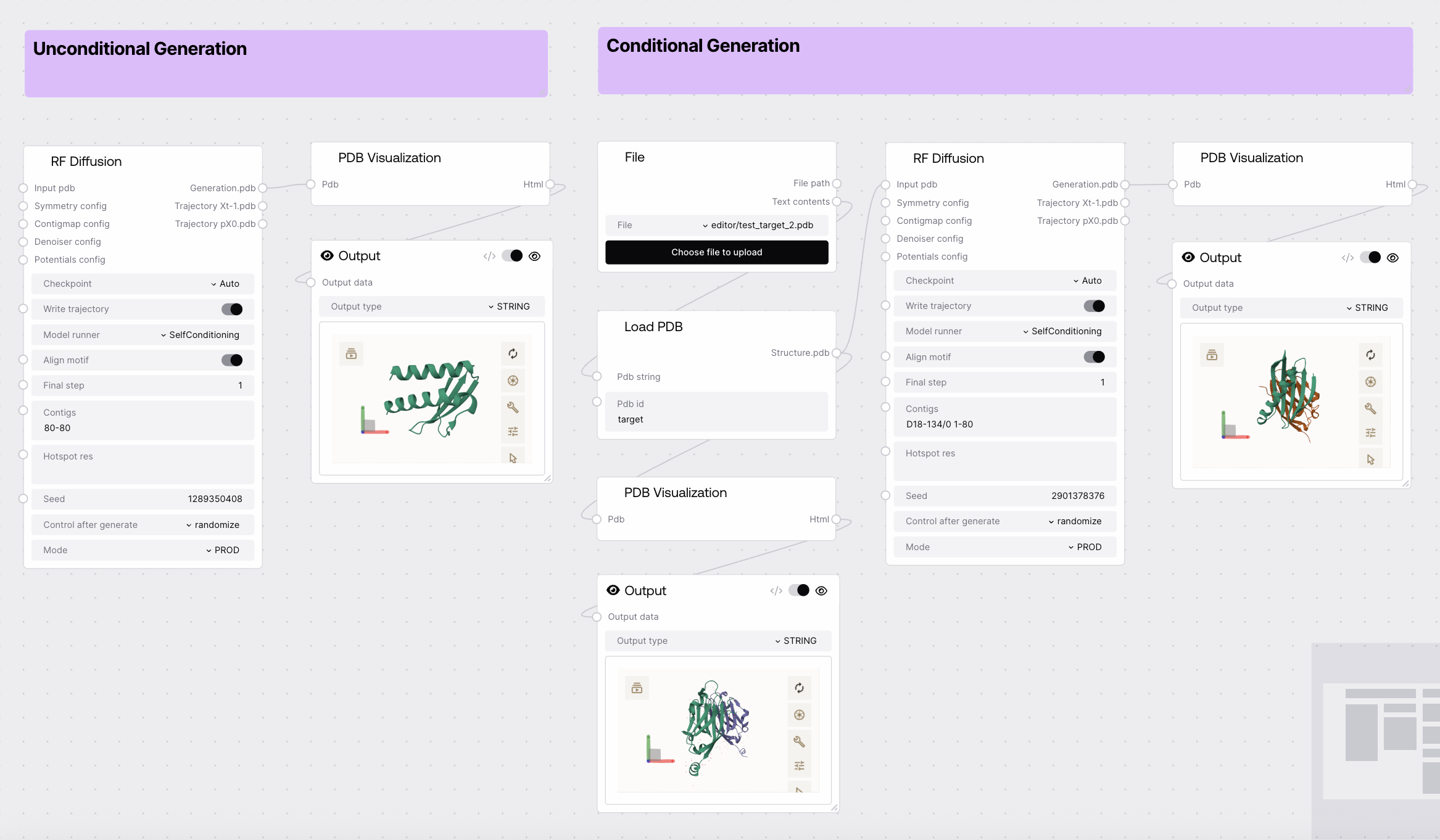
Quick Start
- Select the appropriate checkpoint or use "Auto" for automatic selection.
- Specify the generation mode by setting
contigs (length range for unconditional, motif/gap layout for conditional).
- (Optional) Provide an input PDB for conditional generation.
- Adjust other parameters as needed (e.g.,
write_trajectory, final_step).
- Run the node to generate protein structures.
Setup Guide
1. Unconditional Generation
- Set
contigs to a length range (e.g., 100-100).
- Leave
input_pdb empty.
- Configure other parameters as desired.
2. Conditional Generation (Motif Scaffolding)
- Set
contigs to specify fixed motifs and flexible gaps (e.g., 5-15/A10-25/30-40/0 B1-100).
- Provide an
input_pdb containing the motif.
- (Optional) Connect additional configuration nodes for symmetry, contigmap, denoiser, or potentials.
Basic Usage
Unconditional Generation
- Generate novel protein backbones of specified length.
- Use for de novo protein design.
Motif Scaffolding
- Scaffold a fixed motif within a generated backbone.
- Specify complex layouts with flexible regions and chain breaks.
Configuration
| Field |
Description |
Type |
Example |
| checkpoint |
Checkpoint name or "Auto" for automatic selection. |
COMBO |
Auto |
| write_trajectory |
Whether to include the protein sample trajectory. |
BOOLEAN |
True |
| model_runner |
Sampler name. |
COMBO |
SelfConditioning |
| align_motif |
Align the model's prediction of the motif to the input motif. |
BOOLEAN |
True |
| final_step |
Step to stop reverse diffusion (higher = faster, less accurate). |
INT |
1 |
| contigs |
Sequence layout: length range (unconditional) or motif/gap layout (conditional). |
STRING |
100-100 or 5-15/A10-25 |
| hotspot_res |
Residues on the target protein to involve in inference (comma-separated). |
STRING |
A30,A33,A34 |
| seed |
Base seed for randomness. |
INT |
42 |
| mode |
Node execution mode: MOCK, PROD, or TEST. |
COMBO |
PROD |
| Field |
Description |
Type |
Example |
| input_pdb |
Initial PDB to start generation from (for conditional generation). |
PDB |
|
| symmetry_config |
Additional symmetry configuration (from RFDiffusionSymmetryConfig node). |
JSON |
|
| contigmap_config |
Additional contigmap configuration (from RFDiffusionContigmapConfig node). |
JSON |
|
| denoiser_config |
Additional denoiser configuration (from RFDiffusionDenoiserConfig node). |
JSON |
|
| potentials_config |
Additional potentials configuration (from RFDiffusionPotentialsConfig node). |
JSON |
|
Outputs
| Field |
Description |
Type |
Example |
| generation.pdb |
Generation result. |
PDB |
|
| trajectory_Xt-1.pdb |
Xt-1 trajectory PDB (if write_trajectory is True). |
PDB |
|
| trajectory_pX0.pdb |
pX0 trajectory PDB (if write_trajectory is True). |
PDB |
|
Best Practices
- For unconditional generation, specify only a length range in
contigs and leave input_pdb empty.
- For motif scaffolding, provide both a motif-containing
input_pdb and a detailed contigs string.
Advanced Configuration
- Use configuration nodes for symmetry, contigmap, denoiser, or potentials to fine-tune generation.
- Start with
MOCK or TEST mode to validate setup before running full PROD jobs.
Troubleshooting
Common Issues
- Error: Non-empty
contigs must be passed: Ensure you specify a valid length range or motif/gap layout.
- Error: For unconditional sampling, expected
contigs to specify only length range: Remove motifs or chain breaks from contigs.
- Error: For conditional sampling, expected contigs to specify at least one fixed region: Add a motif region (e.g.,
A10-25) to contigs.
Need Help?
