Haddock¶
Predicts docking of candidate proteins against a target protein using the HADDOCK protocol. Returns the best docking pose, all docked models, and a table of docking scores.
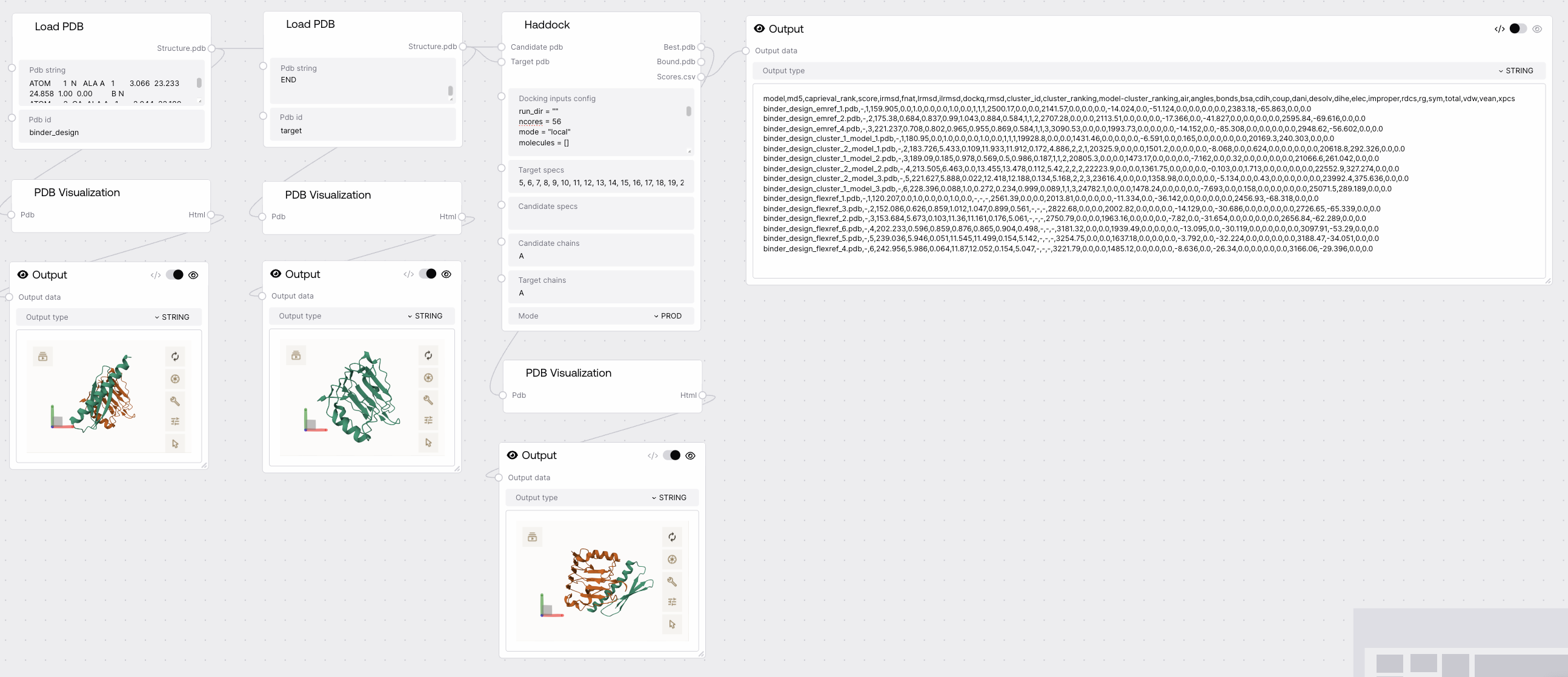
Quick Start¶
- Prepare candidate and target PDB structures using Load PDB node.
- (Optional) Adjust docking configuration parameters as needed.
- Connect candidate and target PDBs to the Haddock node and run docking.
Setup Guide¶
1. Prepare Input Structures¶
- Use the Load PDB node to load candidate and target protein structures.
- Ensure only one target PDB is provided (batching not supported for target).
2. Configure Docking Parameters¶
- Optionally edit the TOML configuration for HADDOCK.
- Specify interface residues and chains if required.
3. Run Docking¶
- Connect all required inputs and execute the node.
- Review outputs: best docking, all dockings, and scores table.
Basic Usage¶
Standard Docking¶
- Dock a single candidate protein against a single target protein.
- Specify interface residues and chains for precise docking.
- Use default or custom HADDOCK configuration.
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| candidate_pdb | Candidate protein structure to dock. | PDB | {"design_0": "..."} |
| target_pdb | Target protein structure to dock against (must be a single PDB). | PDB | {"target": "..."} |
| docking_inputs_config | HADDOCK configuration parameters in TOML format. | STRING | see below |
| target_specs | Residue numbers in the target protein defining the binding interface. | STRING | 1,2,3 |
| candidate_specs | Residue numbers in the candidate protein defining the binding interface. | STRING | 10,11,12 |
| candidate_chains | Chains in the candidate protein to consider for docking. | STRING | A,B |
| target_chains | Chains in the target protein to consider for docking. | STRING | A |
| mode | Execution mode: MOCK, PROD, or TEST. | STRING | PROD |
Optional Inputs¶
None
Outputs¶
| Field | Description | Example |
|---|---|---|
| best.pdb | Best of the predicted dockings. | {"design_0": "..."} |
| bound.pdb | All predicted dockings. | {"design_0_model1": "..."} |
| scores.csv | Docking scores table (caprieval statistics). | CSV string |
Best Practices¶
Input Preparation¶
- Use high-quality, properly formatted PDB files for both candidate and target.
- Ensure only one target PDB is provided per run.
Docking Configuration¶
- Adjust interface residues and chains for more accurate docking.
- Use TEST mode for quick validation with minimal sampling.
Troubleshooting¶
Common Issues¶
- Multiple target PDBs provided: Only one target protein is supported. Use Load PDB to select a single target.
- Docking fails or returns empty results: Check that input PDBs are valid and interface residues/chains are correctly specified.
Need Help?¶
- HADDOCK documentation
- Contact support for further assistance.