Evaluate Unconditional¶
Runs unconditional evaluation pipeline on the outputs of Alphafold or Colabfold models. Selects a folding that is most similar to the generation and returns a scores table.
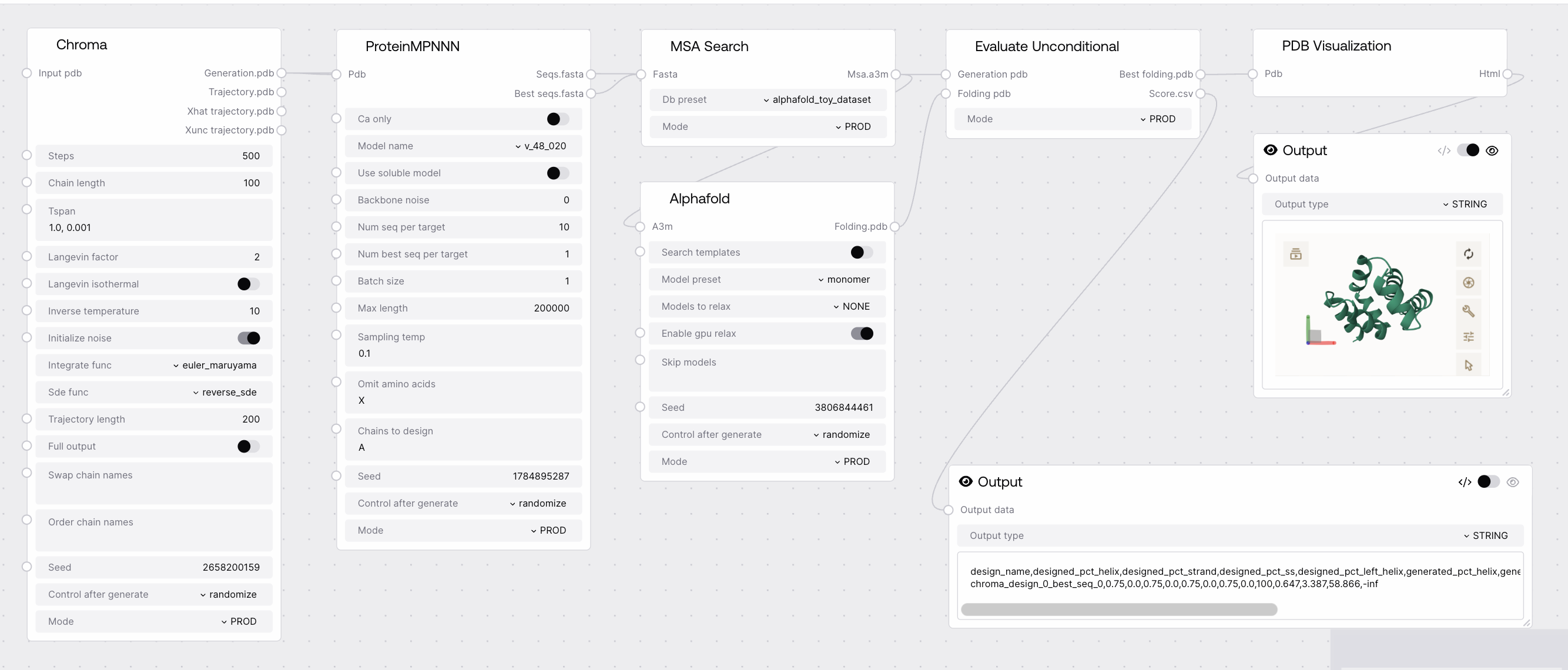
Quick Start¶
- Connect generated proteins (PDB) to
generation_pdbinput. - Connect foldings (PDB) to
folding_pdbinput. - Run the node to obtain the best folding and evaluation scores.
Setup Guide¶
1. Prepare Inputs¶
- Generate proteins using a diffusion or design node.
- Obtain foldings from Alphafold or Colabfold nodes.
2. Connect and Run¶
- Connect the generated proteins to
generation_pdb. - Connect the foldings to
folding_pdb. - Execute the node to receive evaluation results.
Basic Usage¶
Unconditional Evaluation¶
- Compare generated proteins to predicted foldings.
- Select the best matching folding for each design.
- Output a CSV table with evaluation metrics (pst, rmsd, pae, plddt).
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| generation_pdb | Generated proteins to evaluate. | PDB | {"design_0": "...PDB..."} |
| folding_pdb | Foldings to evaluate proteins against. | PDB | {"design_0_ranked_0": "...PDB..."} |
Optional Inputs¶
None
Outputs¶
| Field | Description | Example |
|---|---|---|
| best_folding.pdb | Best of the predicted foldings. | {"design_0": "...PDB..."} |
| score.csv | Scores of the foldings (pst, rmsd, pae, plddt). | "design_0,0.95,1.2,10.5,85.0" |
Best Practices¶
Input Preparation¶
- Ensure that
generation_pdbandfolding_pdbhave matching or corresponding IDs for accurate evaluation. - Use outputs from supported generation and folding nodes for compatibility.
Efficient Evaluation¶
- For large batches, consider running in TEST mode to limit evaluation to the first structure for faster feedback.
Troubleshooting¶
Common Issues¶
- Mismatched IDs: Ensure that the IDs in
generation_pdbandfolding_pdbcorrespond to the same designs. - Empty Outputs: Check that both inputs contain valid PDB data.
Need Help?¶
- Contact support for further assistance.