Evaluate Prot Param¶
Runs protein parameters evaluation on the provided batch of proteins, calculating various sequence and structure properties at specified pH values.
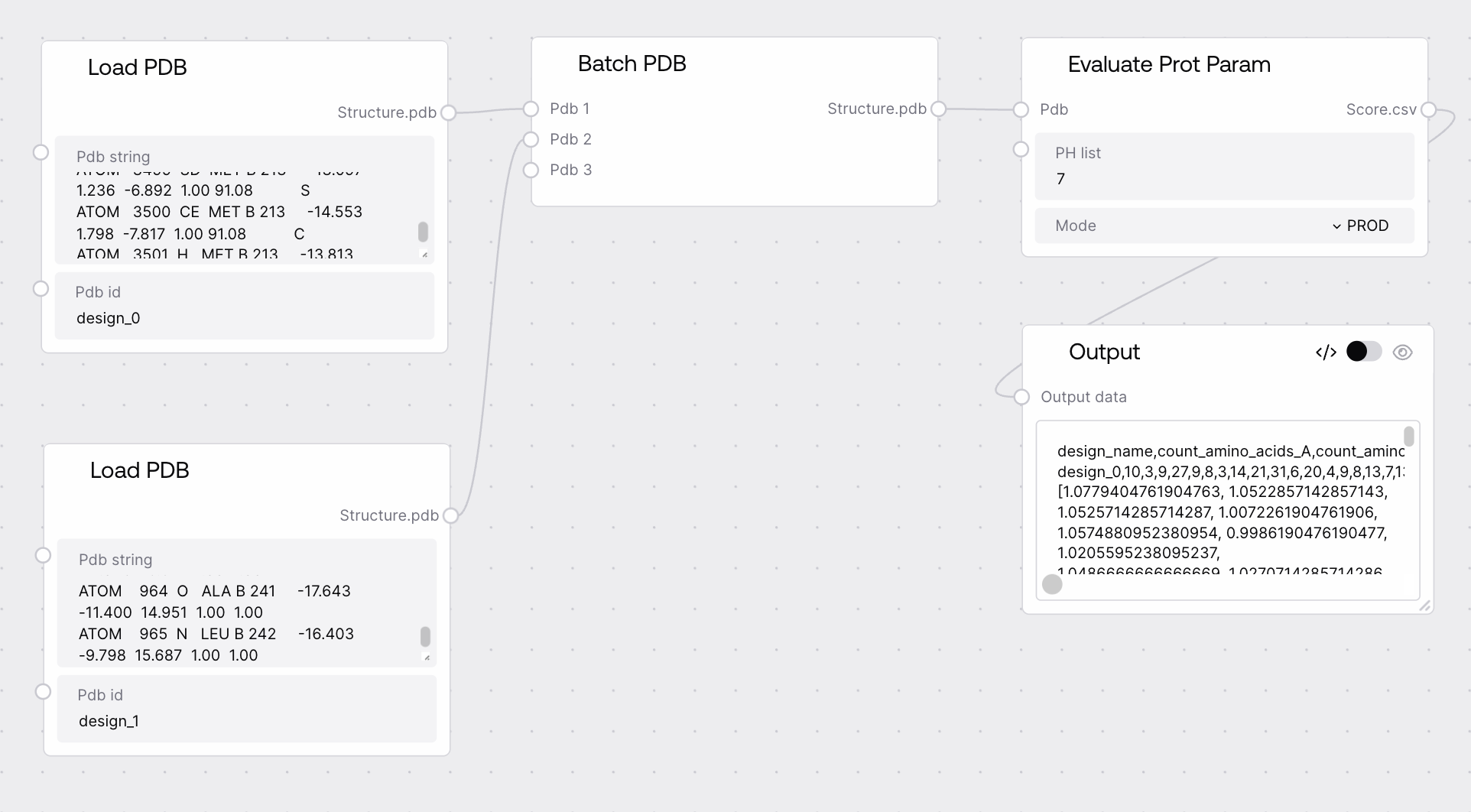
Quick Start¶
- Connect a PDB batch to the node's input.
- Specify a comma-separated list of pH values (e.g.,
7,5.5,8). - Run the node to obtain a CSV table of protein parameters.
Setup Guide¶
1. Prepare Input Data¶
- Generate or load a batch of protein structures in PDB format.
- Ensure each PDB has a unique identifier.
2. Configure Node¶
- Set the
pH_listfield to the desired pH values (comma-separated, e.g.,7,5.5,8). - Connect the PDB input.
3. Run Evaluation¶
- Execute the node to compute parameters for each structure at each specified pH.
- Download or inspect the resulting CSV output.
Basic Usage¶
Batch Protein Parameter Evaluation¶
- Evaluate isoelectric point, charge, and other sequence/structure properties for multiple proteins at different pH values.
- Use in workflows to compare designed proteins or validate generated structures.
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| pdb | Structures to evaluate. | PDB | {"design_0": "...PDB..."} |
| pH_list | Comma-separated list of float pHs to calculate protein charge. | STRING | 7,5.5,8 |
Optional Inputs¶
None
Outputs¶
| Field | Description | Example |
|---|---|---|
| score.csv | Scores of the foldings (various protein/sequence params) | CSV |
Best Practices¶
Input Preparation¶
- Use unique IDs for each PDB to avoid confusion in batch outputs.
- Provide realistic pH values relevant to your experimental or biological context.
Workflow Integration¶
- Combine with diversity or functional evaluation nodes for comprehensive protein assessment.
- Use in automated pipelines to benchmark multiple designs.
Troubleshooting¶
Common Issues¶
- Empty or malformed PDB input: Ensure all PDBs are valid and properly formatted.
- Incorrect pH format: Use comma-separated floats (e.g.,
7,5.5), not lists or other delimiters.
Need Help?¶
- Contact support for further assistance.