Evaluate Diversity¶
Runs diversity evaluation pipeline on the provided batch of proteins. Clusters structures based on TM-score and outputs a CSV table with cluster indices for each input.
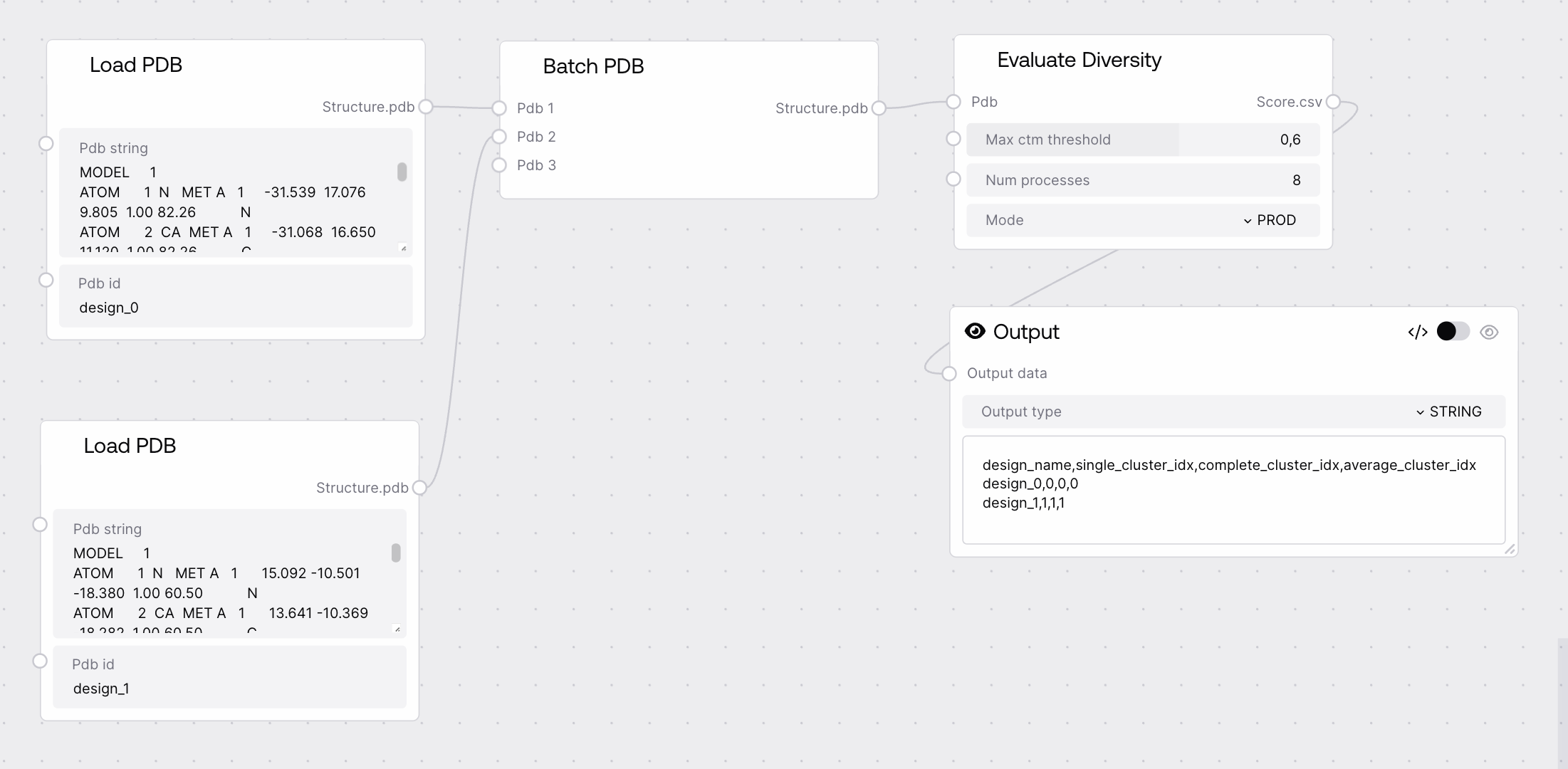
Quick Start¶
- Connect a batch of PDB structures to the node.
- Set the maximum TM-score threshold for clustering (e.g., 0.6).
- Specify the number of processes for parallel evaluation (default: 8).
- Run the node to obtain a CSV with cluster assignments.
Setup Guide¶
1. Prepare Input Structures¶
- Generate or collect a batch of PDB structures to evaluate.
- Ensure each structure has a unique identifier.
2. Configure Diversity Evaluation¶
- Set
max_ctm_thresholdto control cluster separation (lower = stricter clustering). - Adjust
num_processesfor available compute resources.
Basic Usage¶
Batch Diversity Assessment¶
- Cluster a set of generated or designed proteins to assess structural diversity.
- Identify representative structures from each cluster for downstream analysis.
Configuration¶
Required Inputs¶
| Field | Description | Type | Example |
|---|---|---|---|
| pdb | Structures to evaluate. | PDB | {"design_1": "...", "design_2": "..."} |
| max_ctm_threshold | Maximum TM score threshold between clusters. | FLOAT | 0.6 |
| num_processes | Number of processes for parallel evaluation. | INT | 8 |
Optional Inputs¶
None
Outputs¶
| Field | Description | Example |
|---|---|---|
| score.csv | Scores of the pdbs (cluster indices). | CSV file with cluster assignments |
Best Practices¶
Diversity Parameter Tuning¶
- Use a lower
max_ctm_thresholdfor stricter clustering (more clusters, higher diversity sensitivity). - Increase
num_processesto speed up evaluation on large batches if hardware allows.
Workflow Integration¶
- Use after protein generation to select diverse candidates for further analysis.
- Combine with functional evaluation nodes for comprehensive assessment.
Troubleshooting¶
Common Issues¶
- All structures assigned to one cluster: Try lowering
max_ctm_thresholdto increase sensitivity. - Node runs slowly on large batches: Increase
num_processesif possible, or reduce batch size.
Need Help?¶
- Contact support for further assistance.